2012 Prioritizing Sites for Outplanting & Assisted Migration for Strategic Genetic Conservation
Project: Prioritizing sites for outplanting and assisted migration for strategic genetic conservation of whitebark pine
Agency/Forest or Park/District: University of Montana, Division of Biological Sciences, Missoula, MT 59846
Project coordinator: Erin Landguth
Contact: Erin Landguth, erin.landguth@mso.umt.edu, 406.210.9332
Cooperators
Zack Holden and Renate Bush (USFS Region 1); Solomon Dobrowski (Asst. Professor, University of Montana); Mary Mahalovich (Regional Geneticist, USFS Regions 1-4) ; Michael Schwartz (Conservation Genetics Team Leader/Research Ecologist, USFS RMRS); Deborah Mackillop (Research Ecologist, Canadian Forest Service, British Columbia).
Source of funding /amount
FHP: $20,000
Supplemental funding: none
Dates of restoration efforts
2012
Objectives
Reduced Budget and Focus: The project can be reduced to a pilot study on adaptive landscape genetics for WBP for the Inland NW area with expected outcomes: 1) Inland NW maps of current and one future climatic suitability for WBP and 2) Pilot simulations and maps showing locations on the landscape that are important to maintain connectivity and evolutionary resilience across current scenarios, using relative local adaptation, genetic isolation, and demographic vulnerability mapping.
Acres/ha treated
Idaho and Montana
Methods
Climate Envelope Modeling. We will develop high spatial resolution (30m) maps of the bioclimatic distribution of WBP across all areas in and around USFS Region 1, including National Parks and Canada (1 degree north of the US border). A dense network of ~1500 temperature sensors distributed across the region in 2009-2011 will be used to produce high resolution historical and future climate surfaces from General Circulation Model (GCM) outputs, following methods described by Holden et al. (2011). These climate surfaces will give us unprecedented insights into the topoclimatic variation in air temperatures characteristic of the Northern Rockies, including cold air drainage and lapse rate variability. Modeled temperature and downscaled PRISM precipitation surfaces with maps of solar radiation, wind speed and soil depth will be used to develop very high resolution (30m) climatic water balance models using the Penman-Monteith approach. Water balance deficit and minimum temperature maps will then be used with plot data from Forest Inventory and Analysis (FIA), NPS, and the Canadian forest service to produce both current and future bioclimatic suitability models for WBP.
Simulating Genetic Connectivity and Local Adaptation. The recently developed program CDPOP (Landguth and Cushman 2010; Landguth et al. 2011) is the first spatially-explicit, individual-based, evolutionary landscape genetics program that incorporates all factors – mutation, gene flow, genetic drift, and selection – that affect the frequency of an allele in a population. Natural selection is incorporated by imposing differential survival rates defined by local relative fitness values on a landscape. Selection coefficients can vary not only for genotypes, but also in space as functions of local environmental variability. The simulator enables coupling of gene flow (governed by landscape surfaces), with natural selection (governed by selection surfaces).
The ability to couple gene flow that is governed by the underlying landscape with natural selection, which is governed by spatially-explicit selection surfaces, allows for a broad range of applications in adaptive landscape genetics. For example, how does fine-scale environmental variability affect the balance of local adaptation and gene flow? What are the balances between rate of change of the environment, species-specific movement abilities, mutation rates, and selection strengths? Where will resistant traits do the best, or what landscape locations are most vulnerable to genetic loss given projected changes in habitat suitability? This simulator is the basic tool needed to address questions of how spatially and temporally varying environments affect the adaptation of populations to perturbations, such as caused by climate change, anthropogenic changes in landuse and land cover, and innate spatial and temporal dynamics of natural landscapes (see reviews Morris 2011; Neale & Kremer 2011).
WBP Vulnerability Mapping. SDMs of WBP will be used as the landscape surfaces in CDPOP giving estimates of individual locations and habitats. Current and future climatic suitability surfaces will be used as the environmental fitness surfaces in CDPOP controlling natural selection at the fine-scale spatial resolution of interest. Knowledge about particular traits under selection will be linked to their respective environmental locations through increased rate of survival (i.e., increased selection coefficients with respect to the background of non-fit individuals). CDPOP will be used to prioritize areas of greatest concern for WBP on the landscapes with regard to future changing surfaces, by assessing whether or not current traits can locally spread (adapt) fast enough with the changing environment. Specifically, suitability mapping with simulations will identify landscape locations that are spatially isolated and at risk of the quickest extinction. In addition, simulations with transplanted individuals with resistant traits will be conducted to assess outplanting strategies and assisted migration. CDPOP will be used to simulate the change in gene flow and rate of spread for resistant traits across the changing future surfaces, producing knowledge about subpopulation connectivity and structure. The simulations will address how shrinking habitat, individual and genetic isolation, and local adaptation will influence WBP persistence/extinction. By using the genetic information at the individual and fine-scale level, vulnerability maps (both demographics and genetics) will be produced following methods developed by Landguth (in prep.; i.e., population and genetic information concerning diversity within and among subpopulations, as well as temporally).
WBP Interactive Mapping. Our analysis will be made available to the public through the Computational Ecology Laboratory website http://cel.dbs.umt.edu. In addition to the content available on the website, a series of interactive online mapping applications will be available to users (see http://cel.dbs.umt.edu/webapp for a prototype). Our web mapping applications will be intuitive, easy to use, and will be explained by an accompanying series of video tutorials on how to use these maps. The maps will also provide quick visualization capabilities for non-scientific audiences to whom conservation needs are of great importance. In addition to visualization, our interactive maps will also function as a decision support system. This decision support system will provide online geospatial tools to help end-users prioritize areas of conservation concern. These tools will also provide end-users with the ability to download output files from geospatial databases for a user-defined extent viewable in commercial or open-source GIS software packages
Planting? If so, source of seedlings? Resistance? No
Outcome
Inland NW maps of current and one future climatic suitability for WBP
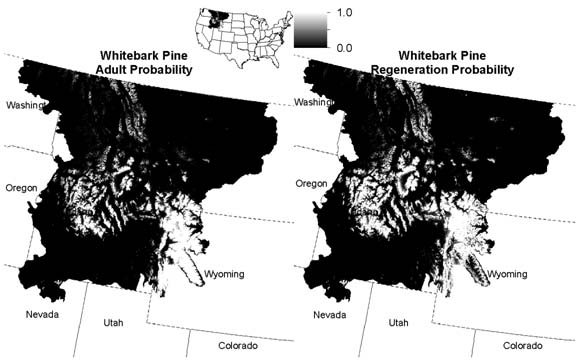
Our initial results include a habitat suitability map of current Whitebark Pine distribution and regeneration across Idaho and Montana (Fig. 1) generated by co-PI Holden.
Figure 1: Whitebark pine habitat suitability maps with adult probability (left panel) and regeneration probability (right panel).
Pilot simulations: simulating genetic connective in current conditions for WBP and local adaption
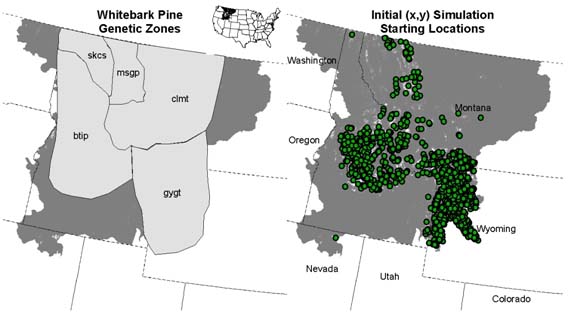
We have held an initial project meeting with staff from USFS Region 1 and Cooperator Mahalovich to identify data needed to parameterize the simulation models. Mahalovich (USDA Forest Service Proceedings RMRS-P-63. 2011) identified provisional seed zones of which five are within our study area (Fig. 2). Although Mahalovich (2011) found very low (FST < 0.026) and little genetic differentiation between zones, these areas and data can provide initial starting locations for individuals and genetic parameters for the simulation model. For exploratory pilot simulations and sensitivity analysis, we have initialized starting simulation (x,y) with 1,000, 5,000, and 10,000 in areas of adult habitat suitability probability of greater than 0.5 (Fig. 2).
Figure 2: Whitebark pine zones of genetic variation identified by Mahalovich (2011) (left panel) and example 1,000 initial starting (x,y) locations for pilot simulations.
For each population (n = 1000 and 5000 individuals) of sexually reproducing individuals distributed randomly within suitable habitat, we modeled spatial changes in allele frequencies for 20 bi-allelic, dominant loci, consisting of one locus under directional selection and 19 neutral loci. We used the spatial regeneration probability surface as a proxy for a spatial selection surface and coded probabilities greater than 0.9 to have a 100% differential mortality with probabilities less than 0.9 having 0% differential mortality. By using this framework, we can test the adaptive potential of offsprings ability for regeneration and thus the conditions in which survival is optimized. We assumed an isolation-by-distance movement and allowed pollen to disperse 10% of the entire range and seed to disperse 75% of the entire study area.
We calculated the genetic diversity (Ho) from the spatially referenced genotypes grouped into genetic neighborhoods generated from the simulations after 100 years. We compared two scenarios in which the influence of the regeneration surface is tied to offspring differential mortality. Figure 3 shows the scenario in which the regeneration niche had no influence over the survival probability of offsprings (Fig. 3 left panel) compared to the regeneration niche influencing the survival probability of offspring (Fig. 3 right panel). Because more regeneration surface exists in the lower right corner of the study area, a relatively genetically robust group of individuals emerge compared to the rest of the study area. However, this results can be dependent on a number of parameters that need further exploration. In particular, dispersal capability, selection or mortality strengths, number and placement of individual locations can all influence the spatial genetic structure that emerges.
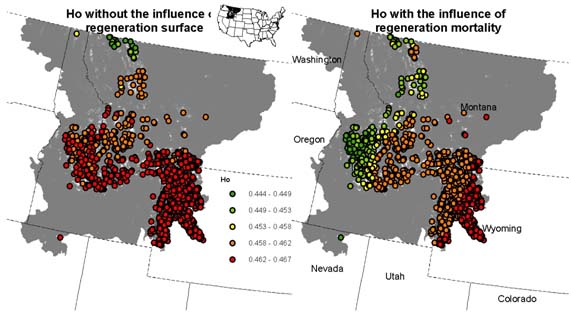 Figure 3: Genetic diversity at spatial referenced genetoypes for 100 years of simulating genetic exchange for individuals in WBP habitat. The right panel includes the influence of differential mortality supplied by the regeneration niche compared to the left panel in which a settling offspring have no survival differences spatially.
Figure 3: Genetic diversity at spatial referenced genetoypes for 100 years of simulating genetic exchange for individuals in WBP habitat. The right panel includes the influence of differential mortality supplied by the regeneration niche compared to the left panel in which a settling offspring have no survival differences spatially.
Monitoring since completion of the project
This project relies entirely on existing and simulated data. The project is still in progress.
Dates
Plans for future monitoring?
No future monitoring is expected.
Will outcome meet goals?
This project is a pilot study which is expected to provide information on the utility of genetic simulation modeling for informing Whitebark management. We expect to meet these goals.
Future actions/follow up?
Inland NW maps of current and one future climatic suitability for WBP
The Whitebark Pine habitat suitability maps for the historic/current distribution of WBP have been completed (Fig. 1). In addition, the future potential habitat suitability maps that are needed for the genetic vulnerability analysis are in preparation. This part of the project is being done by co-PI Holden.
Simulating genetic connective in current conditions for WBP and local adaption
We ran initial pilot simulations to test surface development and model response to neutral versus selection driven loci given offspring settling in a regeneration niche. Although these simulations provide a first glimpse at WBP genetic vulnerability, more sensitivity analysis is needed to assess the potential range of outcomes.
Trees are capable of long-distance gene flow, which can promote adaptive evolution in novel environments by increasing genetic variation for fitness. It is unclear, however, if this can compensate for maladaptive effects of gene flow and for the long-generation times of trees. Recent reviews have suggested that genes can move over spatial scales larger than habitat shifts predicted under climate change within one generation, but differences can occur spatially for example at leading edges versus the cores.
We are planning a more comprehensive study to assess the range of possible outcomes for dispersal strategies within WBP and selection niches. To do this, we will look at numbers and placement of individuals, various gene flow hypotheses (i.e., dispersal mechanisms), and different selection pressures applied via differential mortality across space.
In addition, once the future habitat suitability maps are completed, we will begin genetic simulation analyses to identify the temporal component of ‘how fast can gene flow and selection keep up with changing climate’ and assess potential genetic and demographic vulnerability.